The recent shift in funding policies favoring translational-over-basic research has forced researchers to associate their gene/protein of interest to diseases like cancer. The trend is to check the correlation between the gene of interest and patient survival. The availability of big data sets and improved statistical tools have made it possible, even for basic researchers, to check survival data of their genes. While these advancements do endow us with the ability to make life-saving predictions, not everything that correlates is actually relevant.
Let’s think of it this way, it can be so that you are just a handshake away from shaking hands with Mark Hamill ( Luke Skywalker of Star Wars) but does that mean you two are nearly friends? As baby Yoda would say “nearly friends, you are not”. Causation and correlation have a kinda similar relationship. Especially in complex diseases like cancer, which depends on DNA damage and takes a long time to become pathogenic. A lot of changes are just the by-products of the genomic instability, however, they may never play any significant role in cancer progression. Just like you may never actually shake hands with Mark Hamill, even though you are just a single handshake away from him. Therefore, making research decisions based on correlative data is a high-risk bet, which needs a thorough evaluation to avoid serious setbacks.
While writing my previous blog on Kaplan-Meier plot, a friend of mine directed me to a twitter thread from @JSheltzer, where he brings up the issue of correlation in research. Do check it out as some of the examples are adopted from his tweet. Let’s look at some of the examples where the predictive power of big data betrays us.
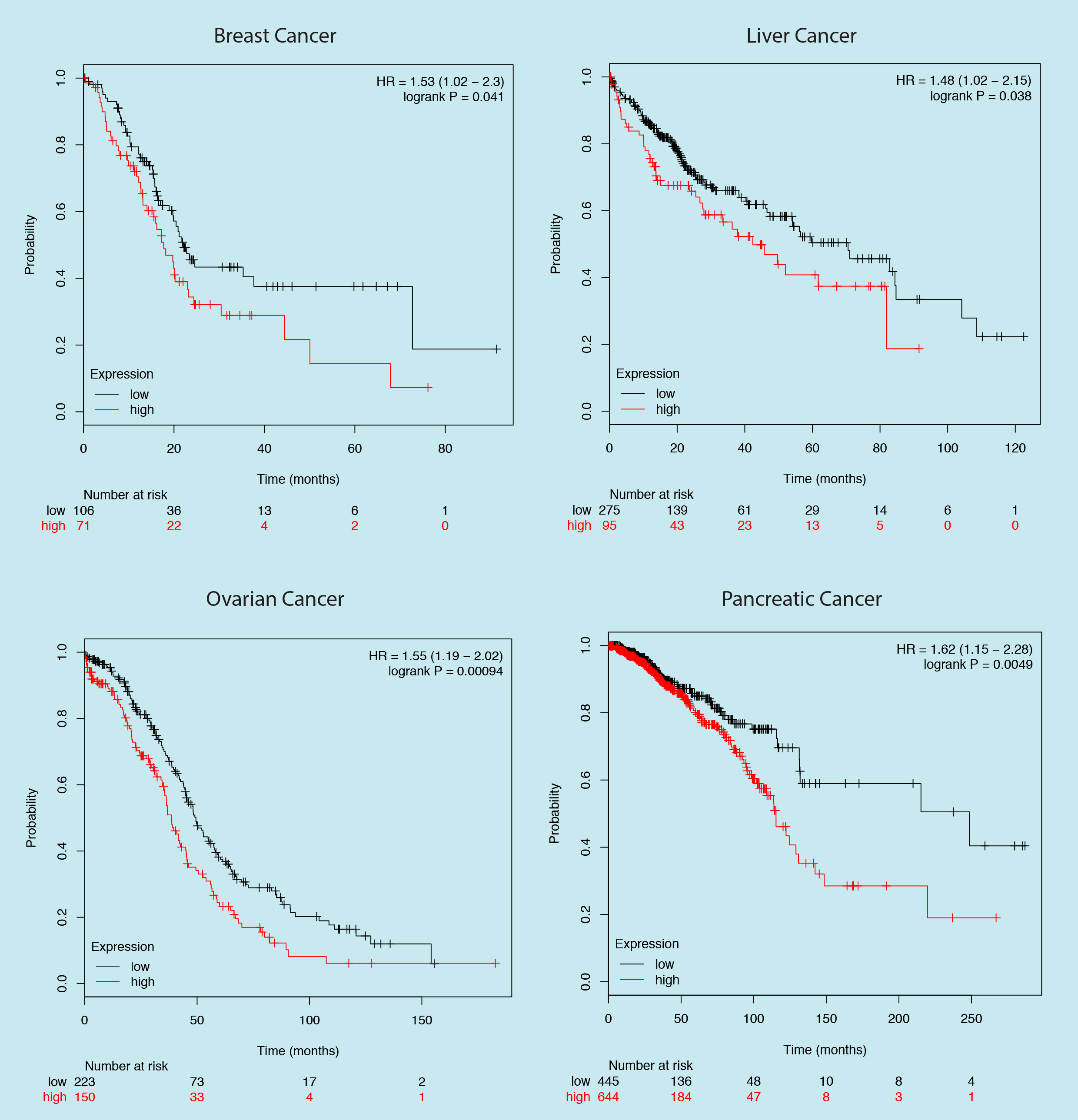
Here is a survival curve plotted for the expression of a gene in four different cancer types, in each case high gene expression is significantly associated with poor prognosis. If you have to make a research decision based on this data would you choose to inhibit the protein?
If your answer is yes, then you just choose to inhibit RB1, a well-known tumor suppressor.
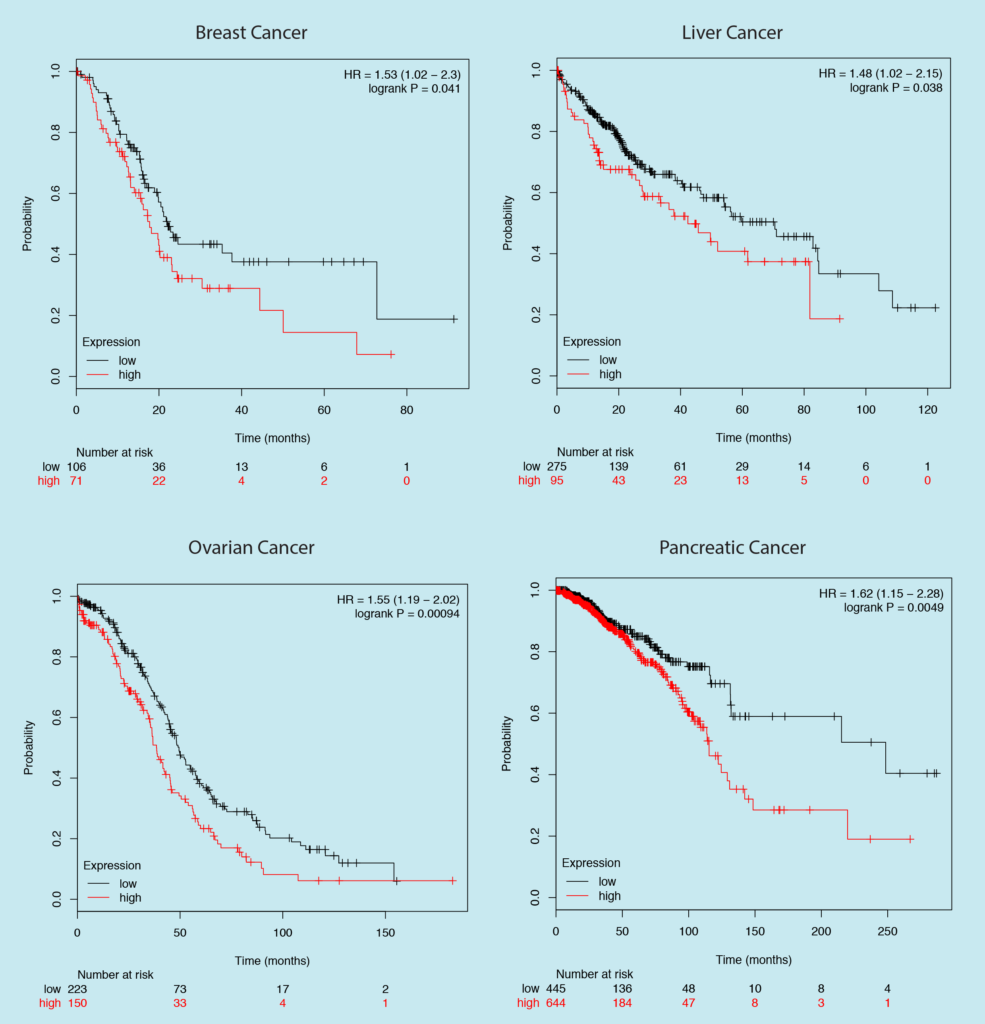
Let’s look at another example, here is a gene whose expression is significantly associated with a better prognosis in four different cancer types. It won’t be a smart idea to target it right?
Too bad, Nobel Prize just slipped out of your hands. The above survival plot is for the expression of PDL1 — an immune checkpoint molecule which has radically changed the cancer therapy.
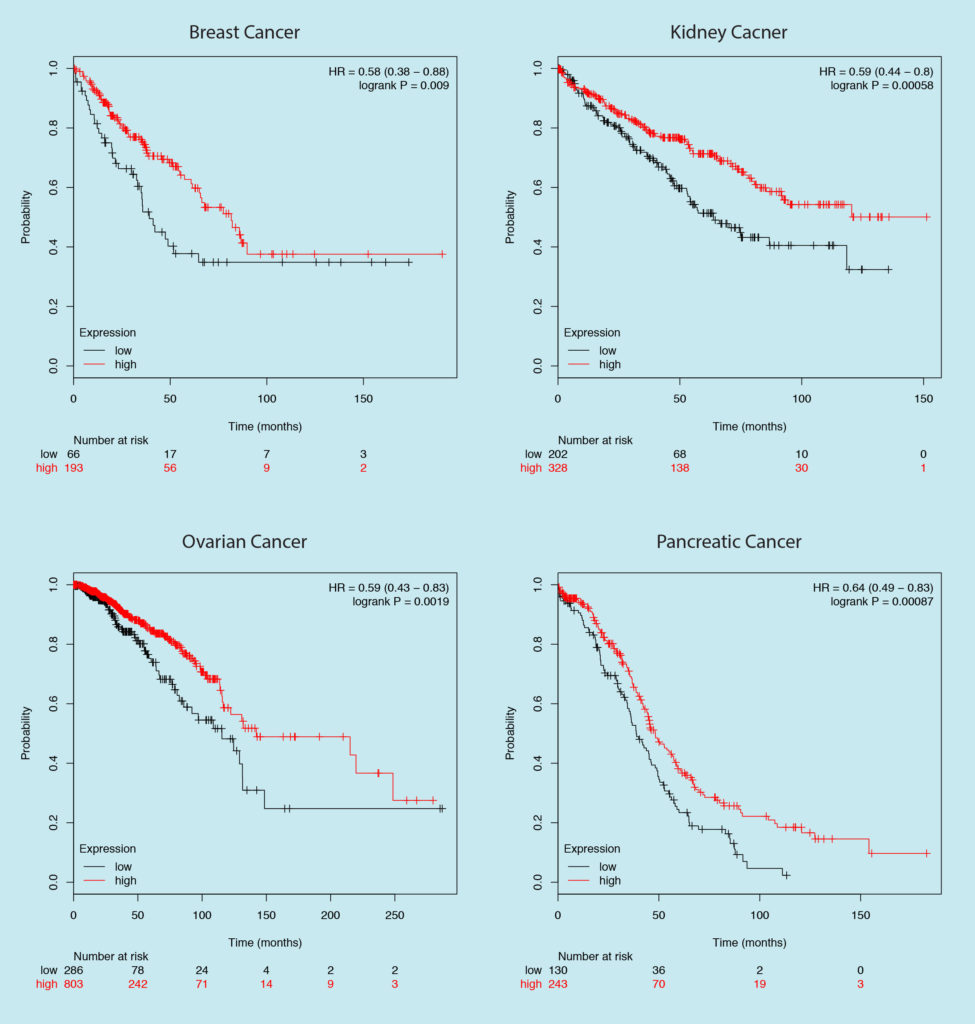
Let’s be a bit more specific, based on the survival curves for the expression of two separate genes in breast cancer shown below, which one would be your choice of the target?
The one on the left is the Estrogen Receptor, a well-established target in breast cancer. If you choose the one on the right then you just decided to target GAPDH — your good old housekeeping gene.
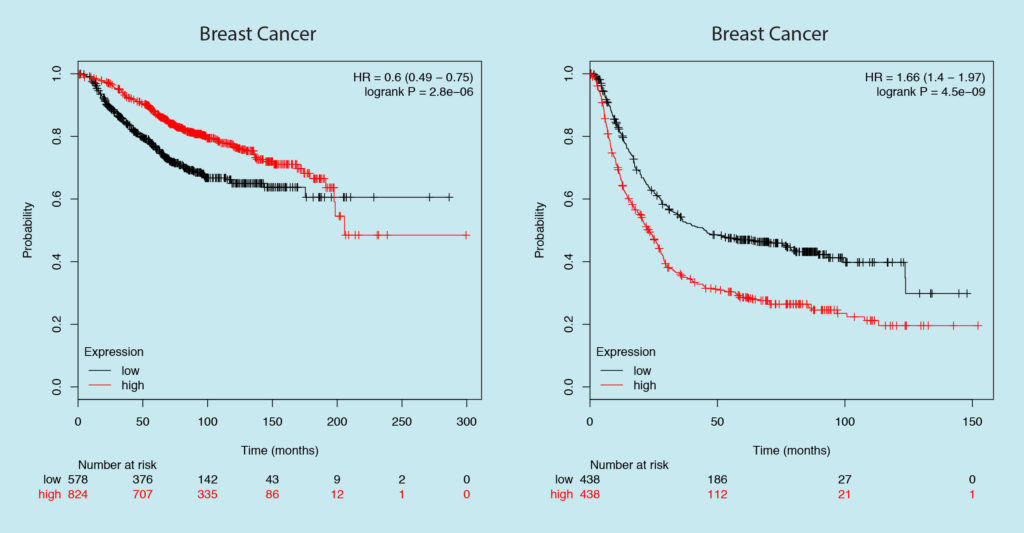
Let’s look at Gastric cancer this time. Which gene would you target, one on the left or one on the right?

If your answer is one on the right then congrats! You targeted GAPDH again. The survival plot on the left is of KRAS — one of the known oncogenes, based on this data KRAS expression seems to improve patient survival in Gastric cancer.
With these examples, I am not trying to imply that big data sets lack predictive power. However, I want to emphasize that at times certain alterations can just be a by-product of genomic instability, which may never play any significant role in cancer progression. In other cases, as in the case of the Estrogen receptor in breast cancer, the good prognosis might be because of the well-established therapy against it. So, correlating expression data with poor prognosis is not strong evidence to justify the importance of the gene as a cancer driver.